ONGOING PROJECTS
- Development of Remote Radiation Detection Imaging System Mounted on a Drone
- High Resolution SPECT using Variable Pinhole Collimator
- Positioning Algorithm for CZT Virtual Frisch-grid Detector
- Multi-purpose Super-resolution Gamma Detector
- Multi-pinhole SPECT
- Low Profile Light Guide using Diffusion Film
- Advanced Reconstruction for Radiation imaging (ARRA)
COMPLETED PROJECTS
- Reconstruction of Dose Distribution in In-beam PET for Carbon Theraphy
- Image Registration for Breast Cancer Study
- High Energy Collimator Design for I-131
- Plasma-Display-Panel based X-ray Detector (PXD)
- CCD based Gamma Camera
- Abdomen Registration for PET/CT and MR images
- Performance Optimization by Patient Dose Analysis and Image Quality Assessment in CT Fluoroscopy
- Cone-beam based system matrix for a pixelated SPECT detector
- Development of Time-of-Flight Method for Improvement of Signal-to-Noise Ratio
Abdomen Registration for PET/CT and MR images
Introduction
The aim of this study is to develop a 3D registration algorithm for PET/CT and MR images acquired from independent PET/CT and MR imaging systems. Combined PET/CT images provide anatomical and functional information and MR images have high resolutions for soft tissues. With the registration technique, the strengths of each modality image can be combined so that we can achieve higher performance in diagnosis and radiotherapy planning. For the case of abdomen registration, the organs are too flexible to be solved by same method because they are non-rigidly transformed by the breath and position of the patient. For these reasons and to get more precise abdominal registration results, we have studied a feature based registration method to overcome the drawback of the conventional method described above. As a method to extract features, independent component analysis (ICA) was explored. To improve the performance of the voxel method, particle swarm optimization (PSO) and GPU based parallel processing was employed.
Materials and Methods
The proposed method consists of two stages: normalized mutual information (NMI) based global matching and independent component analysis (ICA) based refinement.
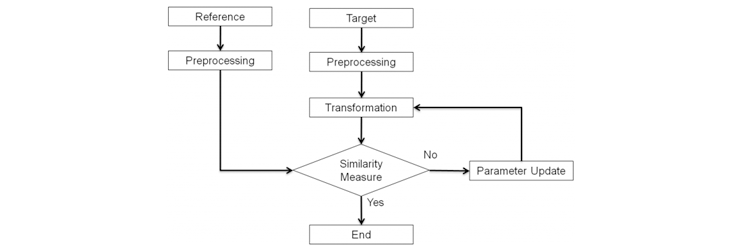
Fig. 1. Block diagram of NMI based global matching.
The goal of the NMI based global matching stage is to roughly match the images with the global information of the CT and MR images. The global registration stage consists of 4 steps (preprocessing, geometrical transformation, similarity measure, and optimization).
In the global matching, the field of view of the CT and MR images are adjusted to the same size in the preprocessing step. Then, the target image is geometrically transformed, and the similarities between the two images are measured with NMI.
The optimization step updates the transformation parameters to efficiently find the best matched parameter set.
We developed a feature based registration model to improve the results of the NMI based global matching. To supplement the limitation of the conventional intensity based method, we explored an additional step which extracts features based on ICA followed by elastic transformation.
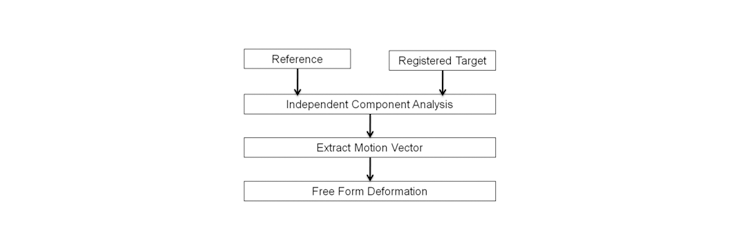
Fig. 2 Block diagram of ICA based refinement.
In this ICA based refinement stage, ICA planes from the windowed image slices were extracted and the similarity between the images was measured to determine the transformation parameters of the control points. Figure 3 illustrates ICA planes from the reference and target slices. In this example, reference 2 (dashed box) best matched the target slice. B-spline based freeform deformation was performed for the geometrical transformation.
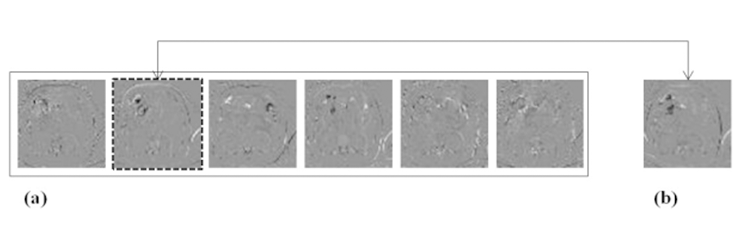
Fig. 3 Extracted feature from reference slices (a) and target slice (b). Dashed box image is the best matching ICA slice.
Results
The proposed algorithm was implemented in a Linux machine with c++ and CUDA technology. To analyze the results easily, we also developed a 3D image viewer which can display the reference PET/CT, target MR and the resulting registered image in a window. The proposed registration algorithm consists of two independent registration methods, NMI based global matching and ICA based refinement. The result image set of each method evaluated independently.
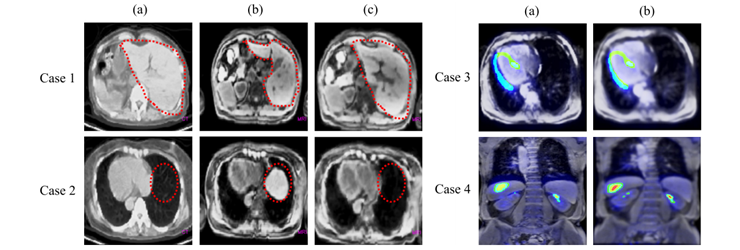
Fig. 4. Case(1) and Case(2). Two cases of global registration result. (a) is reference CT, (b) is target MR and (c) is MR after global registration. Case(3) and Case(4). Two cases of PET/MR fusion image for global registration (a) and refinement (b).
Figure 4 case 1 and case 2 show the NMI based global matching. For the case 1, the shape of liver in CT image (a) and MR image (b) were different before the registration, but after the global matching(c), the size and shape of liver was considerably registered. For the case 2 image, unlike the CT image (a), the liver was shown in MR image (b). In registered MR image(c), the liver is disappeared and the size of heart is quite analogous to CT image.
Figure 4 case 3 and case 4 show the results of ICA based fine adjustment. The case 3 is transverse image of heart area. The wall of left ventricle was left-sided in global registered fusion image (a). After the ICA based refinement, PET/MR image shows that misalignment has been considerably corrected (b). The case 4 is an image set of a patient who has some suspicious region at the lower side of liver. After the refinement process (b), the lesion of ROI is successfully matched in PET/MR fusion image.
Conclusion
The aim of this study is to develop an image registration algorithm for abdominal PET/CT and standalone MR images. We developed a 3D elastic image registration algorithm which consists of NMI based global matching and ICA-based refinement. The NMI based global matching algorithm adopted parameters for the geometrical transformation and NMI for the similarity measurement. For efficient updates of the Affine parameters, a particle swarm optimization method was adopted. In ICA based refinement, ICA feature planes were utilized and matched to determine the motion vectors of 64 control points in a 3D space. Based on the control points, we transformed the target image elastically by B-spline Abased freeform deformation. To supplement the shortcomings of the conventional intensity based method, we developed an additional ICA based refinement step. Performance of this proposed hybrid method has been verified by matching PET/CT and MR images. Moreover, the reliability of this method will enable the algorithm to be utilized for CT-CT registration as well as CT-MR registration in the future. Furthermore, we expect the proposed algorithm will be applied to various combinations of multi-modality imaging applications.
Participating Researchers



